An practical guide of statistical analyses in Neuroscience studies (for beginners)
Published:
Introduction
This notebook aims to provide statistical functions, build dataFrame, and perform statistical analyses. The conditions/groups are push/pull+ healthy/lf-patients/hf-patients/sham-patients + before/after-TMS The hypotheses tested are:
- difference between healthy subjects and patients (includng lf/hf/sham-TMS) in push/pull epochs()
- the regression between CMMN properties (including components-SPMIs and graph properties) and FMA
- clustering analyses on CMMN properties (UE-brunnstrom clusters)
- ANOVA and MANOVA and post-hoc analyses (details are missed)
Section1: fcs & load basic epoch info
from itertools import count
import matplotlib,numpy as np
import matplotlib.cm as cm
class EEGChannel:
'''innitilized by info chs'''
_ids = count(0)
def __init__(self, x, y,name="None"):
self.id = next(self._ids)
self.x = x
self.y = y
self.name = name
def plot(self,ax,center):
circle = plt.Circle((center[0]+self.x, center[1]+self.y), radius=0.2,fill = True)
ax.add_patch(circle)
label = ax.annotate(self.name, xy=(center[0]+self.x, center[1]+self.y), fontsize=18, ha="center")
return ax
class EMGChannel:
'''with polar coordinates as inputs'''
_ids = count(0)
def __init__(self, rho, phi,name="None"):
self.id = next(self._ids)
self.rho = rho
self.phi = phi
self.name = name
self.x = rho * np.cos(phi/180*np.pi)
self.y = rho * np.sin(phi/180*np.pi)
def plot(self,ax,center):
circle = plt.Circle((center[0]+self.x, center[1]+self.y), radius=0.2,fill = True)
ax.add_patch(circle)
label = ax.annotate(self.name, xy=(center[0]+self.x, center[1]+self.y), fontsize=18, ha="center")
return ax
def cartesianC(self):
return(self.x, self.y)
def polarC(self):
return(self.rho,self.phi)
# EMG Topograph_basis
def plotTopoEMG(EMGs,ax,center=[7,0]):
for i in range(2):
circle = plt.Circle((center[0],center[1]), 2*(i+1),fill = False)
ax.add_artist(circle)
for EMG in EMGs:
ax = EMG.plot(ax,center)
return ax
def plotTopoEEG(EEGs,ax,center=[-5,0],radius=6):
circle = plt.Circle((center[0],center[1]),radius,fill = False)
ax.plot()
ax.plot([center[0]-0.5,center[0]],[radius+center[1],radius+center[1]+1])
ax.plot([center[0]+0.5,center[0]],[radius+center[1],radius+center[1]+1])
ax.add_artist(circle)
for EEG in EEGs:
ax = EEG.plot(ax,center)
return ax
def c_map(data):
'''
data should be numpy array (adj_m, for example)
'''
minima = np.min(data[np.nonzero(data)])
maxima = np.max(data)
if minima==maxima:
minima-=0.2
maxima+=0.2
norm = matplotlib.colors.Normalize(vmin=minima,vmax=maxima,clip=True)
# mapper = cm.ScalarMappable(norm=norm,cmap=cm.viridis)
mapper = cm.ScalarMappable(norm=norm,cmap="bwr")
return mapper
def c_map_bwr(data):
'''
data should be numpy array (adj_m, for example)
'''
minima = np.min(data[np.nonzero(data)])
maxima = np.max(data)
if minima==maxima:
minima-=0.2
maxima+=0.2
boundary = (np.abs(maxima)+np.abs(minima))/2
norm = matplotlib.colors.Normalize(vmin=-boundary,vmax=boundary,clip=True)
# mapper = cm.ScalarMappable(norm=norm,cmap=cm.viridis)
mapper = cm.ScalarMappable(norm=norm,cmap="bwr")
return mapper
def graph_plot(adj_m,fig,ax,ignore_zero=True,center_EEG=[-5,0],center_EMG=[7,0]):
mapper = c_map(adj_m)
if ignore_zero==True:
for ix,iy in np.ndindex(adj_m.shape):
if -0.1<adj_m[ix,iy]<0.1:
pass
else:
if ix>31:
if iy>31:
ax.plot([EMGs[ix-32].x+center_EMG[0], EMGs[iy-32].x+center_EMG[0]],
[EMGs[ix-32].y+center_EMG[1], EMGs[iy-32].y+center_EMG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
else:
ax.plot([EMGs[ix-32].x+center_EMG[0], EEGs[iy].x+center_EEG[0]],
[EMGs[ix-32].y+center_EMG[1], EEGs[iy].y+center_EEG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
else:
if iy>31:
ax.plot([EEGs[ix].x+center_EEG[0], EMGs[iy-32].x+center_EMG[0]],
[EEGs[ix].y+center_EEG[1], EMGs[iy-32].y+center_EMG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
else:
ax.plot([EEGs[ix].x+center_EEG[0], EEGs[iy].x+center_EEG[0]],
[EEGs[ix].y+center_EEG[1], EEGs[iy].y+center_EEG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
fig.colorbar(mapper,ax=ax)
return fig,ax
def graph_plot_fixMapper(adj_m,adj_m_fix,fig,ax,ignore_zero=True,center_EEG=[-5,0],center_EMG=[7,0]):
# the colorbar of the output plot only depends on adj_m_fix
mapper = c_map_bwr(adj_m_fix)
if ignore_zero==True:
for ix,iy in np.ndindex(adj_m.shape):
if -0.1<adj_m[ix,iy]<0.1:
pass
else:
if ix>31:
if iy>31:
ax.plot([EMGs[ix-32].x+center_EMG[0], EMGs[iy-32].x+center_EMG[0]],
[EMGs[ix-32].y+center_EMG[1], EMGs[iy-32].y+center_EMG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
else:
ax.plot([EMGs[ix-32].x+center_EMG[0], EEGs[iy].x+center_EEG[0]],
[EMGs[ix-32].y+center_EMG[1], EEGs[iy].y+center_EEG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
else:
if iy>31:
ax.plot([EEGs[ix].x+center_EEG[0], EMGs[iy-32].x+center_EMG[0]],
[EEGs[ix].y+center_EEG[1], EMGs[iy-32].y+center_EMG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
else:
ax.plot([EEGs[ix].x+center_EEG[0], EEGs[iy].x+center_EEG[0]],
[EEGs[ix].y+center_EEG[1], EEGs[iy].y+center_EEG[1]],
color = mapper.to_rgba(adj_m[ix,iy]))
fig.colorbar(mapper,ax=ax)
return fig,ax
# setup
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy
from statannot import add_stat_annotation
from itertools import count
import os, mne, warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
# calculate the Cohen's d between two samples
from numpy.random import randn
from numpy.random import seed
from numpy import mean
from numpy import var
from math import sqrt
# function to calculate Cohen's d for independent samples
def cohend(d1, d2):
n1, n2 = len(d1), len(d2)
s1, s2 = var(d1, ddof=1), var(d2, ddof=1)
s = sqrt(((n1 - 1) * s1 + (n2 - 1) * s2) / (n1 + n2 - 2))
u1, u2 = mean(d1), mean(d2)
return (u1 - u2) / s
chs_list_preImage_lParetic = ['Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'Fz','Cz','Pz','Oz',
'Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_rParetic = ['Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'Fz','Cz','Pz','Oz',
'Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
################## Dataset Loading ##################
data_dir = 'E:/masterSJTU/MultiEEGEMG_stroke/'
iVC_results_dir = os.path.join(data_dir,'results','iVC')
if not os.path.exists(iVC_results_dir):
os.makedirs(iVC_results_dir)
# data_dir = 'E:/important file/Projects/CM-graph/sample_data' #PC dir
subj_idx = '15'
contraction_type = 'iMC'
session_idx = 's01'
subjInfo_fName = os.path.join(data_dir,'subj_info.txt')
subjInfo = pd.read_csv(subjInfo_fName, skiprows=0,
sep = ',',engine = 'python')
affected_h = subjInfo[subjInfo['subj_idx']==int(subj_idx)]['affected_h'].values[0]
epochs_hybrid_dir = os.path.join(data_dir,'subj'+subj_idx,'epochs_hybrid')
epochs_hybrid_fName = os.path.join(data_dir,'subj'+subj_idx,'epochs_hybrid',
'subj'+subj_idx+'_'+contraction_type+'_'+session_idx+'_hybrid_epo.fif')
epochs_hybrid = mne.read_epochs(epochs_hybrid_fName,preload=True)
EMGs=[EMGChannel(2,150,'emg1'),EMGChannel(2,170,'emg2'),EMGChannel(2, 240,'emg3'),EMGChannel(2,350,'emg4'),
EMGChannel(2, 80,'emg5'),EMGChannel(4,140,'emg6'),EMGChannel(4, 310,'emg7'),
EMGChannel(4,90,'emg8')]
EEGs = [EEGChannel(x=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][0]*50,
y=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][1]*50,
name=ch) for ch in chs_list_image[:32]]
Reading E:/masterSJTU/MultiEEGEMG_stroke/subj15\epochs_hybrid\subj15_iMC_s01_hybrid_epo.fif ...
Found the data of interest:
t = 0.00 ... 2000.00 ms
0 CTF compensation matrices available
45 matching events found
Applying baseline correction (mode: mean)
Not setting metadata
0 projection items activated
Section2: df construction
import os, pandas as pd, numpy as np
# definition
n_chs = 40
cV_ratio = 5
n_SPMIs = int((n_chs*n_chs-n_chs)/2)
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
patient_subjs = ['3','6','8','11','31','33','40','22','23','26','28','29','38','39','41','37','36','12','16']
healthy_subjs = ['13','14','15','17','18','19','21']
data_dir = r'E:\masterSJTU\MultiEEGEMG_stroke'
# data_dir = 'E:/important file/Projects/CM-graph/sample_data' #PC dir
global_results_dir = os.path.join(data_dir,'results')
contraction_type = 'iVC'
if not os.path.exists(global_results_dir):
os.makedirs(global_results_dir)
subj_infofName = os.path.join(data_dir,'subj_info.txt')
subj_info_df = pd.read_csv(subj_infofName, delimiter = ",")
# df generation
df_colNames = ['subj_id','subj_group','FMA','contraction_group']
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
df_colNames.append('SPMI'+'_'+ch1_name+'_'+ch2_name)
df_gProperties_fName = os.path.join(data_dir,'subj3','temp','subj3iVC_gProperties_push_be4TMS.csv')
df_gProperties = pd.read_csv(df_gProperties_fName, skiprows=0,sep = ',',engine = 'python')
df_colNames+=df_gProperties.columns.tolist()
df_data = None
for subj in patient_subjs:
for contraction_group in ['push','pull']:
temp_dir = os.path.join(data_dir,'subj'+subj,'temp')
SPMI_withoutD_z_fName = os.path.join(temp_dir,contraction_type+'_z_SPMI_'+contraction_group+'_be4TMS')
SPMI_withoutD_z = np.load(SPMI_withoutD_z_fName+'.npy')
df_gProperties_fName = os.path.join(temp_dir,'subj'+subj+contraction_type+'_gProperties_'+contraction_group+'_be4TMS.csv')
df_gProperties = pd.read_csv(df_gProperties_fName, skiprows=0,sep = ',',engine = 'python')
df_SPMI_data = []
FMA = subj_info_df.loc[subj_info_df['subj_idx']==int(subj),'FMA'].values[0]
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
df_SPMI_data.append(SPMI_withoutD_z[ch1_idx,ch2_idx])
if df_data is None:
df_data = np.array([subj,'patient',FMA,contraction_group]+df_SPMI_data+df_gProperties.iloc[0].values.tolist())
else:
df_data = np.vstack((df_data,np.array([subj,'patient',FMA,contraction_group]+df_SPMI_data+df_gProperties.iloc[0].values.tolist())))
for subj in healthy_subjs:
for contraction_group in ['push','pull']:
temp_dir = os.path.join(data_dir,'subj'+subj,'temp')
SPMI_withoutD_z_fName = os.path.join(temp_dir,contraction_type+'_z_SPMI_'+contraction_group+'_be4TMS')
SPMI_withoutD_z = np.load(SPMI_withoutD_z_fName+'.npy')
df_gProperties_fName = os.path.join(temp_dir,'subj'+subj+contraction_type+'_gProperties_'+contraction_group+'_be4TMS.csv')
df_gProperties = pd.read_csv(df_gProperties_fName, skiprows=0,sep = ',',engine = 'python')
df_SPMI_data = []
FMA = subj_info_df.loc[subj_info_df['subj_idx']==int(subj),'FMA'].values[0]
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
df_SPMI_data.append(SPMI_withoutD_z[ch1_idx,ch2_idx])
if df_data is None:
df_data = np.array([subj,'healthy',FMA,contraction_group]+df_SPMI_data+df_gProperties.iloc[0].values.tolist())
else:
df_data = np.vstack((df_data,np.array([subj,'healthy',FMA,contraction_group]+df_SPMI_data+df_gProperties.iloc[0].values.tolist())))
df = pd.DataFrame (df_data, columns = df_colNames)
# manipulate data
df['FMA']=df['FMA'].astype(float)
df.iloc[:,4:4+n_SPMIs] = df.iloc[:,4:4+n_SPMIs].astype(float)
measuresContaining_inf = ['_closeness_vitality','_closeness_vitality_weight','_cost_closeness_vitality_weight']
for ch in chs_list_image:
for measure in measuresContaining_inf:
df[ch+measure] = df[ch+measure].astype(float)
df[ch+measure].replace(-np.inf,np.nan, inplace = True)
df[ch+measure].replace(np.nan,-df[ch+measure].mean()*cV_ratio, inplace = True)
df_fName = os.path.join(global_results_dir,'iVC','iVC_be4TMS_df.csv')
df.to_csv(df_fName)
Section 3: group difference (healthy vs patients_be4TMS) during pushing and pulling.
Section 3-1: anova_SPMIs: subj_group and contraction_group as independent variables
from bioinfokit.analys import stat
pd.set_option('display.expand_frame_repr', False)
n_chs = 40
alpha=.01
cons_sigDiff = []
res = stat()
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
res_var = 'SPMI_'+ch1_name+'_'+ch2_name
res.anova_stat(df=df, res_var=res_var, anova_model=res_var+' ~ C(contraction_group)*C(subj_group)',ss_typ=3)
sigDiff = [True if PR<alpha else False for PR in res.anova_summary['PR(>F)'].tolist()]
if True in sigDiff[1:]:
cons_sigDiff.append('SPMI_'+ch1_name+'_'+ch2_name)
print(ch1_name+'_'+ch2_name)
print(res.anova_summary)
print('\n')
FC5i_emg7
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.441531 0.441531 0.851944 0.360620
C(contraction_group) 1.0 0.052691 0.052691 0.101669 0.751220
C(subj_group) 1.0 3.829877 3.829877 7.389838 0.009100
C(contraction_group):C(subj_group) 1.0 0.799676 0.799676 1.542993 0.220208
Residual 48.0 24.876608 0.518263 NaN NaN
CP5i_P3i
df sum_sq mean_sq F PR(>F)
Intercept 1.0 546.242738 546.242738 27.281337 0.000004
C(contraction_group) 1.0 1.861847 1.861847 0.092987 0.761731
C(subj_group) 1.0 189.380907 189.380907 9.458367 0.003464
C(contraction_group):C(subj_group) 1.0 3.199880 3.199880 0.159814 0.691102
Residual 48.0 961.083805 20.022579 NaN NaN
CP5i_emg7
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.230313 0.230313 0.517084 0.475575
C(contraction_group) 1.0 0.269040 0.269040 0.604031 0.440858
C(subj_group) 1.0 3.356947 3.356947 7.536794 0.008481
C(contraction_group):C(subj_group) 1.0 0.014565 0.014565 0.032699 0.857263
Residual 48.0 21.379573 0.445408 NaN NaN
O1i_PO3c
df sum_sq mean_sq F PR(>F)
Intercept 1.0 388.114280 388.114280 39.739512 8.594169e-08
C(contraction_group) 1.0 22.912381 22.912381 2.346028 1.321675e-01
C(subj_group) 1.0 103.182268 103.182268 10.564963 2.109636e-03
C(contraction_group):C(subj_group) 1.0 17.855648 17.855648 1.828262 1.826696e-01
Residual 48.0 468.789987 9.766458 NaN NaN
Cz_emg3
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.992398 0.992398 3.256988 0.077395
C(contraction_group) 1.0 0.029442 0.029442 0.096627 0.757262
C(subj_group) 1.0 3.518057 3.518057 11.546038 0.001373
C(contraction_group):C(subj_group) 1.0 0.071678 0.071678 0.235244 0.629869
Residual 48.0 14.625514 0.304698 NaN NaN
Pz_PO3c
df sum_sq mean_sq F PR(>F)
Intercept 1.0 223.870742 223.870742 30.812939 0.000001
C(contraction_group) 1.0 0.000171 0.000171 0.000024 0.996150
C(subj_group) 1.0 56.977905 56.977905 7.842279 0.007333
C(contraction_group):C(subj_group) 1.0 0.150360 0.150360 0.020695 0.886215
Residual 48.0 348.742959 7.265478 NaN NaN
Pz_O1c
df sum_sq mean_sq F PR(>F)
Intercept 1.0 296.164646 296.164646 37.745261 1.511944e-07
C(contraction_group) 1.0 0.154453 0.154453 0.019685 8.890087e-01
C(subj_group) 1.0 83.674398 83.674398 10.664041 2.019270e-03
C(contraction_group):C(subj_group) 1.0 0.176546 0.176546 0.022500 8.813924e-01
Residual 48.0 376.627494 7.846406 NaN NaN
F3c_emg2
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.929862 1.929862 7.665084 0.007977
C(contraction_group) 1.0 0.347687 0.347687 1.380955 0.245735
C(subj_group) 1.0 2.816901 2.816901 11.188254 0.001604
C(contraction_group):C(subj_group) 1.0 0.442735 0.442735 1.758470 0.191091
Residual 48.0 12.085107 0.251773 NaN NaN
T8c_emg1
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.416524 1.416524 8.987323 0.004297
C(contraction_group) 1.0 0.000879 0.000879 0.005575 0.940790
C(subj_group) 1.0 1.586762 1.586762 10.067422 0.002632
C(contraction_group):C(subj_group) 1.0 0.003049 0.003049 0.019345 0.889965
Residual 48.0 7.565452 0.157614 NaN NaN
T8c_emg7
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.129381 0.129381 0.238419 0.627574
C(contraction_group) 1.0 0.187718 0.187718 0.345921 0.559188
C(subj_group) 1.0 4.346094 4.346094 8.008849 0.006777
C(contraction_group):C(subj_group) 1.0 0.722499 0.722499 1.331398 0.254270
Residual 48.0 26.047755 0.542662 NaN NaN
PO3c_O1c
df sum_sq mean_sq F PR(>F)
Intercept 1.0 904.924185 904.924185 33.855088 4.741203e-07
C(contraction_group) 1.0 0.391985 0.391985 0.014665 9.041181e-01
C(subj_group) 1.0 232.517554 232.517554 8.698963 4.908212e-03
C(contraction_group):C(subj_group) 1.0 0.071786 0.071786 0.002686 9.588845e-01
Residual 48.0 1283.008373 26.729341 NaN NaN
emg3_emg5
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.395210 0.395210 0.094226 0.760199
C(contraction_group) 1.0 38.408973 38.408973 9.157522 0.003974
C(subj_group) 1.0 0.369037 0.369037 0.087986 0.768033
C(contraction_group):C(subj_group) 1.0 17.556139 17.556139 4.185760 0.046265
Residual 48.0 201.324179 4.194254 NaN NaN
emg4_emg5
df sum_sq mean_sq F PR(>F)
Intercept 1.0 3.514889 3.514889 2.277745 0.137798
C(contraction_group) 1.0 12.619950 12.619950 8.178075 0.006258
C(subj_group) 1.0 0.238863 0.238863 0.154790 0.695742
C(contraction_group):C(subj_group) 1.0 8.144604 8.144604 5.277928 0.026001
Residual 48.0 74.070926 1.543144 NaN NaN
emg5_emg6
df sum_sq mean_sq F PR(>F)
Intercept 1.0 37.763148 37.763148 13.808449 0.000528
C(contraction_group) 1.0 11.472046 11.472046 4.194861 0.046038
C(subj_group) 1.0 37.938409 37.938409 13.872534 0.000514
C(contraction_group):C(subj_group) 1.0 12.933282 12.933282 4.729176 0.034615
Residual 48.0 131.269713 2.734786 NaN NaN
emg5_emg7
df sum_sq mean_sq F PR(>F)
Intercept 1.0 106.092092 106.092092 14.882442 0.000341
C(contraction_group) 1.0 53.066727 53.066727 7.444122 0.008866
C(subj_group) 1.0 81.340388 81.340388 11.410309 0.001457
C(contraction_group):C(subj_group) 1.0 43.465125 43.465125 6.097223 0.017149
Residual 48.0 342.176404 7.128675 NaN NaN
emg5_emg8
df sum_sq mean_sq F PR(>F)
Intercept 1.0 14.135787 14.135787 10.733318 0.001959
C(contraction_group) 1.0 5.996139 5.996139 4.552875 0.038001
C(subj_group) 1.0 10.414482 10.414482 7.907727 0.007109
C(contraction_group):C(subj_group) 1.0 3.423998 3.423998 2.599845 0.113431
Residual 48.0 63.216034 1.317001 NaN NaN
Section 3-2: tukey HSD_SPMIs
pd.set_option('display.expand_frame_repr', False)
for con_sigDiff in cons_sigDiff:
print(con_sigDiff)
res.tukey_hsd(df=df, res_var=con_sigDiff, xfac_var='contraction_group', anova_model=con_sigDiff+' ~ C(contraction_group)*C(subj_group)')
print(res.tukey_summary)
res.tukey_hsd(df=df, res_var=con_sigDiff, xfac_var='subj_group', anova_model=con_sigDiff+' ~ C(contraction_group)*C(subj_group)')
print(res.tukey_summary)
res.tukey_hsd(df=df, res_var=con_sigDiff, xfac_var=['contraction_group','subj_group'], anova_model=con_sigDiff+' ~ C(contraction_group)*C(subj_group)')
print(res.tukey_summary)
print('\n')
SPMI_FC5i_emg7
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.285917 -0.115538 0.687371 2.025121 0.158631
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.585695 0.133158 1.038232 3.680154 0.012283
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.306117 -0.541049 1.153284 1.360089 0.747640
1 (push, patient) (pull, patient) 0.436459 -0.185192 1.058109 2.642681 0.255085
2 (push, patient) (pull, healthy) 0.428815 -0.418352 1.275982 1.905236 0.534257
3 (push, healthy) (pull, patient) 0.742576 -0.104591 1.589743 3.299287 0.104827
4 (push, healthy) (pull, healthy) 0.122697 -0.901478 1.146872 0.450930 0.900000
5 (pull, patient) (pull, healthy) 0.865273 0.018106 1.712440 3.844434 0.043646
SPMI_CP5i_P3i
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.088025 -2.407268 2.583317 0.100307 0.9
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 5.525298 2.712497 8.338099 5.585536 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 4.966040 -0.299636 10.231715 3.549803 0.071152
1 (push, patient) (pull, patient) 0.389164 -3.474785 4.253113 0.379096 0.900000
2 (push, patient) (pull, healthy) 5.695393 0.429718 10.961068 4.071157 0.029355
3 (push, healthy) (pull, patient) 5.355203 0.089528 10.620879 3.827984 0.044884
4 (push, healthy) (pull, healthy) 0.729353 -5.636538 7.095244 0.431248 0.900000
5 (pull, patient) (pull, healthy) 6.084557 0.818882 11.350232 4.349337 0.017625
SPMI_CP5i_emg7
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.332397 -0.039773 0.704566 2.539595 0.078827
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.772359 0.352834 1.191884 5.234913 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.734628 -0.050739 1.519996 3.520813 0.074525
1 (push, patient) (pull, patient) 0.352713 -0.223589 0.929015 2.303666 0.373284
2 (push, patient) (pull, healthy) 0.457376 -0.327991 1.242744 2.192043 0.417892
3 (push, healthy) (pull, patient) 1.087342 0.301974 1.872709 5.211242 0.003160
4 (push, healthy) (pull, healthy) 0.277252 -0.672211 1.226715 1.099119 0.849790
5 (pull, patient) (pull, healthy) 0.810090 0.024722 1.595457 3.882472 0.040890
SPMI_O1i_PO3c
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.627763 -1.114967 2.370493 1.024268 0.472418
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 3.170116 1.205637 5.134596 4.588554 0.002143
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 1.849023 -1.828561 5.526607 1.892465 0.539257
1 (push, patient) (pull, patient) 0.083595 -2.615014 2.782204 0.116598 0.900000
2 (push, patient) (pull, healthy) 4.407615 0.730031 8.085199 4.511170 0.012954
3 (push, healthy) (pull, patient) 1.932618 -1.744966 5.610202 1.978024 0.505765
4 (push, healthy) (pull, healthy) 2.558592 -1.887390 7.004575 2.166115 0.428480
5 (pull, patient) (pull, healthy) 4.491210 0.813626 8.168794 4.596730 0.010980
SPMI_Cz_emg3
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.030618 -0.277202 0.338438 0.28283 0.846746
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.745598 0.398611 1.092586 6.109971 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.661896 0.012321 1.311470 3.835383 0.044323
1 (push, patient) (pull, patient) 0.075689 -0.400969 0.552346 0.597684 0.900000
2 (push, patient) (pull, healthy) 0.753613 0.104038 1.403187 4.366841 0.017053
3 (push, healthy) (pull, patient) 0.737584 0.088010 1.387159 4.273963 0.020286
4 (push, healthy) (pull, healthy) 0.091717 -0.693580 0.877014 0.439606 0.900000
5 (pull, patient) (pull, healthy) 0.829301 0.179727 1.478876 4.805421 0.007265
SPMI_Pz_PO3c
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.170194 -1.332926 1.673314 0.321958 0.826746
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 3.216214 1.521833 4.910596 5.397368 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 3.094984 -0.076965 6.266933 3.672657 0.058240
1 (push, patient) (pull, patient) 0.235472 -2.092102 2.563046 0.380789 0.900000
2 (push, patient) (pull, healthy) 3.101972 -0.069976 6.273921 3.680950 0.057446
3 (push, healthy) (pull, patient) 3.330456 0.158507 6.502405 3.952080 0.036240
4 (push, healthy) (pull, healthy) 0.006989 -3.827710 3.841688 0.006860 0.900000
5 (pull, patient) (pull, healthy) 3.337445 0.165496 6.509394 3.960373 0.035715
SPMI_Pz_O1c
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.018077 -1.54398 1.580134 0.032907 0.9
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 3.913065 2.152247 5.673883 6.319035 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 3.781701 0.485380 7.078022 4.318232 0.018679
1 (push, patient) (pull, patient) 0.052657 -2.366181 2.471495 0.081940 0.900000
2 (push, patient) (pull, healthy) 3.991771 0.695451 7.288092 4.558106 0.011832
3 (push, healthy) (pull, patient) 3.834358 0.538037 7.130679 4.378360 0.016685
4 (push, healthy) (pull, healthy) 0.210070 -3.774987 4.195128 0.198417 0.900000
5 (pull, patient) (pull, healthy) 4.044428 0.748108 7.340749 4.618234 0.010527
SPMI_F3c_emg2
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.011143 -0.268669 0.290955 0.113236 0.9
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.534047 0.218631 0.849463 4.814419 0.001346
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.326021 -0.264450 0.916492 2.078235 0.464705
1 (push, patient) (pull, patient) 0.100871 -0.332416 0.534158 0.876272 0.900000
2 (push, patient) (pull, healthy) 0.641202 0.050731 1.231673 4.087374 0.028515
3 (push, healthy) (pull, patient) 0.426892 -0.163579 1.017363 2.721243 0.231810
4 (push, healthy) (pull, healthy) 0.315181 -0.398664 1.029026 1.661900 0.629504
5 (pull, patient) (pull, healthy) 0.742073 0.151602 1.332544 4.730381 0.008440
SPMI_T8c_emg1
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.009386 -0.212004 0.230776 0.120551 0.9
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.574214 0.324654 0.823775 6.542537 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.591477 0.124290 1.058665 4.765357 0.007871
1 (push, patient) (pull, patient) 0.018682 -0.324140 0.361503 0.205114 0.900000
2 (push, patient) (pull, healthy) 0.575632 0.108445 1.042820 4.637699 0.010132
3 (push, healthy) (pull, patient) 0.572796 0.105609 1.039983 4.614845 0.010596
4 (push, healthy) (pull, healthy) 0.015845 -0.548957 0.580647 0.105595 0.900000
5 (pull, patient) (pull, healthy) 0.556951 0.089764 1.024138 4.487187 0.013567
SPMI_T8c_emg7
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.619986 0.20919 1.030781 4.291447 0.003882
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.656 0.192933 1.119066 4.028174 0.006452
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.390255 -0.476624 1.257134 1.694487 0.616748
1 (push, patient) (pull, patient) 0.763079 0.126963 1.399194 4.515248 0.012853
2 (push, patient) (pull, healthy) 0.158665 -0.708214 1.025545 0.688925 0.900000
3 (push, healthy) (pull, patient) 1.153334 0.286455 2.020213 5.007773 0.004821
4 (push, healthy) (pull, healthy) 0.231590 -0.816416 1.279596 0.831771 0.900000
5 (pull, patient) (pull, healthy) 0.921744 0.054865 1.788623 4.002212 0.033183
SPMI_PO3c_O1c
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.457084 -2.425989 3.340157 0.450805 0.760882
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 6.825765 3.575841 10.075689 5.97209 0.001
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 6.909531 0.825545 12.993516 4.274729 0.020257
1 (push, patient) (pull, patient) 0.502189 -3.962236 4.966614 0.423399 0.900000
2 (push, patient) (pull, healthy) 7.244188 1.160203 13.328174 4.481772 0.013710
3 (push, healthy) (pull, patient) 6.407342 0.323356 12.491328 3.964039 0.035486
4 (push, healthy) (pull, healthy) 0.334658 -7.020522 7.689838 0.171260 0.900000
5 (pull, patient) (pull, healthy) 6.742000 0.658014 12.825985 4.171082 0.024507
SPMI_emg3_emg5
group1 group2 Diff Lower Upper q-value p-value
0 push pull 1.398134 0.256075 2.540194 3.481034 0.01748
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 1.578561 0.291182 2.865939 3.486607 0.017312
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 2.888528 0.478503 5.298552 4.511317 0.012950
1 (push, patient) (pull, patient) 0.692768 -1.075707 2.461242 1.474474 0.702865
2 (push, patient) (pull, healthy) 0.424174 -1.985850 2.834198 0.662477 0.900000
3 (push, healthy) (pull, patient) 3.581295 1.171271 5.991319 5.593285 0.001393
4 (push, healthy) (pull, healthy) 3.312702 0.399124 6.226279 4.279608 0.020075
5 (pull, patient) (pull, healthy) 0.268594 -2.141431 2.678618 0.419491 0.900000
SPMI_emg4_emg5
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.594827 -0.097904 1.287559 2.4416 0.090696
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.676148 -0.104728 1.457024 2.462111 0.088094
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 1.568386 0.106555 3.030218 4.038349 0.031127
1 (push, patient) (pull, patient) 0.114392 -0.958300 1.187083 0.401391 0.900000
2 (push, patient) (pull, healthy) 0.330482 -1.131350 1.792313 0.850939 0.900000
3 (push, healthy) (pull, patient) 1.682778 0.220946 3.144609 4.332890 0.018176
4 (push, healthy) (pull, healthy) 1.898868 0.131600 3.666136 4.044274 0.030799
5 (pull, patient) (pull, healthy) 0.216090 -1.245741 1.677922 0.556399 0.900000
SPMI_emg5_emg6
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.167174 -0.755022 1.089369 0.515457 0.727832
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 1.598984 0.559445 2.638523 4.373727 0.0033
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.474638 -1.471420 2.420695 0.918025 0.900000
1 (push, patient) (pull, patient) 0.438244 -0.989772 1.866260 1.155130 0.827865
2 (push, patient) (pull, healthy) 2.285087 0.339029 4.231145 4.419725 0.015431
3 (push, healthy) (pull, patient) 0.912881 -1.033176 2.858939 1.765659 0.588890
4 (push, healthy) (pull, healthy) 1.810450 -0.542220 4.163119 2.896502 0.185051
5 (pull, patient) (pull, healthy) 2.723331 0.777273 4.669389 5.267359 0.002808
SPMI_emg5_emg7
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.881335 -0.607567 2.370236 1.683151 0.239833
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 1.92644 0.248086 3.604794 3.263775 0.025361
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.134742 -3.007202 3.276686 0.161418 0.900000
1 (push, patient) (pull, patient) 0.228533 -2.077024 2.534089 0.373096 0.900000
2 (push, patient) (pull, healthy) 3.759089 0.617145 6.901034 4.503316 0.013154
3 (push, healthy) (pull, patient) 0.093791 -3.048154 3.235735 0.112359 0.900000
4 (push, healthy) (pull, healthy) 3.893831 0.095406 7.692257 3.858529 0.042604
5 (pull, patient) (pull, healthy) 3.987622 0.845678 7.129566 4.777093 0.007688
SPMI_emg5_emg8
group1 group2 Diff Lower Upper q-value p-value
0 push pull 0.463368 -0.176594 1.103331 2.058826 0.151954
group1 group2 Diff Lower Upper q-value p-value
0 patient healthy 0.848342 0.126949 1.569735 3.343853 0.022149
group1 group2 Diff Lower Upper q-value p-value
0 (push, patient) (push, healthy) 0.269830 -1.080647 1.620307 0.752058 0.900000
1 (push, patient) (pull, patient) 0.151861 -0.839117 1.142840 0.576808 0.900000
2 (push, patient) (pull, healthy) 1.578716 0.228239 2.929193 4.400126 0.016016
3 (push, healthy) (pull, patient) 0.117968 -1.232508 1.468445 0.328796 0.900000
4 (push, healthy) (pull, healthy) 1.308886 -0.323761 2.941533 3.017573 0.157236
5 (pull, patient) (pull, healthy) 1.426855 0.076378 2.777331 3.976865 0.034700
Section 3-3: SPMI_bp - groupDiff
for con_sigDiff in cons_sigDiff:
plt.figure()
ax = sns.boxplot(x="contraction_group", y=con_sigDiff, hue="subj_group", data=df, palette="Set3")
# res.tukey_hsd(df=df, res_var=con_sigDiff, xfac_var='contraction_group', anova_model=con_sigDiff+' ~ C(contraction_group)*C(subj_group)')
# res.tukey_summary['p-value'].values[0]
fig = ax.get_figure()
fig.set_rasterized(True)
fig.savefig(os.path.join(iVC_results_dir,con_sigDiff+'_ANOVA_bp.eps'))
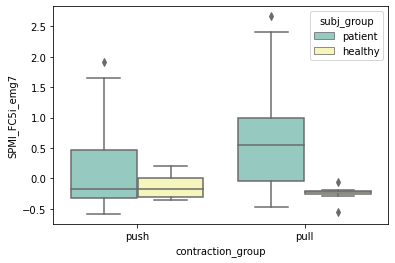
















Section 3-4: interaction Effect
from statsmodels.graphics.factorplots import interaction_plot
alpha = .05
for con_sigDiff in cons_sigDiff:
res.anova_stat(df=df, res_var=con_sigDiff, anova_model=con_sigDiff+' ~ C(contraction_group)*C(subj_group)',ss_typ=3)
# print( res.anova_summary['PR(>F)'][3])
if res.anova_summary['PR(>F)'][3]<alpha:
fig = interaction_plot(x=df['contraction_group'], trace=df['subj_group'], response=df[con_sigDiff],colors=['#4c061d','#d17a22'])
fig.savefig(os.path.join(iVC_results_dir,con_sigDiff+'_ANOVA_profileP.eps'))
plt.show()
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.

The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.

The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.

The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.

Section 3-4: graph properties analyses
from bioinfokit.analys import stat
pd.set_option('display.expand_frame_repr', False)
gProperties= ['radius','diameter', 'density', 's_metric', 'transitivity', 'local_efficiency', 'global_efficiency',
'sw_omega', 'node_connectivity_wholeG','average_shortestPathLength_weight',
'wiener_idx', 'wiener_idx_weight', 'size','size_weight','assortativityCoefficient',
'cost_size_weight', 'cost_wiener_index_weight','cost_average_shortestP_length','cost_average_shortestP_length_weight',
'mean_clusteringC', 'g_cliqueNumber', 'g_numberOfMaxClique','non_randomness','relative_nonRandomness',
'wiener_idx_minST_weight', 'wiener_idx_maxST_weight','size_minST_weight','size_maxST_weight']
gProperties_local = ['eigenvector_centrality','closeness_centrality','degree_centrality','betweenness_centrality','clusteringC_weight','closeness_vitality',
'closeness_vitality_weight','ecc_weight','cost_ecc_weight','cost_betweenness_centrality','cost_eigenvector_centrality',
'cost_closeness_centrality', 'cost_closeness_vitality_weight']
for ch in chs_list_image:
gProperties+=[ch+'_'+gProperty_local for gProperty_local in gProperties_local]
alpha=.05
gProperties_sigDiff = []
res = stat()
for gProperty in gProperties:
res_var = gProperty
df[gProperties] = df[gProperties].astype(float)
res.anova_stat(df=df, res_var=res_var, anova_model=res_var+' ~ C(contraction_group)*C(subj_group)',ss_typ=3)
sigDiff = [True if PR<alpha else False for PR in res.anova_summary['PR(>F)'].tolist()]
if True in sigDiff[1:]:
gProperties_sigDiff.append(gProperty)
print(gProperty)
print(res.anova_summary)
print('\n')
sw_omega
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.114231 0.114231 11.327823 0.001510
C(contraction_group) 1.0 0.065420 0.065420 6.487393 0.014125
C(subj_group) 1.0 0.005956 0.005956 0.590653 0.445932
C(contraction_group):C(subj_group) 1.0 0.021945 0.021945 2.176242 0.146686
Residual 48.0 0.484038 0.010084 NaN NaN
wiener_idx
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.096752e+07 1.096752e+07 187.084528 3.522585e-18
C(contraction_group) 1.0 3.159006e+05 3.159006e+05 5.388649 2.456042e-02
C(subj_group) 1.0 2.255250e+04 2.255250e+04 0.384702 5.380313e-01
C(contraction_group):C(subj_group) 1.0 1.461715e+05 1.461715e+05 2.493401 1.208927e-01
Residual 48.0 2.813920e+06 5.862334e+04 NaN NaN
cost_average_shortestP_length
df sum_sq mean_sq F PR(>F)
Intercept 1.0 17.928206 17.928206 166.023554 3.391861e-17
C(contraction_group) 1.0 0.617292 0.617292 5.716415 2.077689e-02
C(subj_group) 1.0 0.039448 0.039448 0.365305 5.484211e-01
C(contraction_group):C(subj_group) 1.0 0.246829 0.246829 2.285748 1.371235e-01
Residual 48.0 5.183324 0.107986 NaN NaN
F7i_degree_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 2.778623 2.778623 133.070657 1.924401e-15
C(contraction_group) 1.0 0.000751 0.000751 0.035984 8.503474e-01
C(subj_group) 1.0 0.110977 0.110977 5.314779 2.551143e-02
C(contraction_group):C(subj_group) 1.0 0.006676 0.006676 0.319713 5.744139e-01
Residual 48.0 1.002279 0.020881 NaN NaN
FC5i_cost_closeness_vitality_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 9.631906e+05 9.631906e+05 0.417609 0.521210
C(contraction_group) 1.0 1.007313e+07 1.007313e+07 4.367390 0.041956
C(subj_group) 1.0 9.305374e+04 9.305374e+04 0.040345 0.841656
C(contraction_group):C(subj_group) 1.0 4.239610e+06 4.239610e+06 1.838161 0.181511
Residual 48.0 1.107092e+08 2.306441e+06 NaN NaN
P3i_cost_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.147522 0.147522 9.065951 0.004144
C(contraction_group) 1.0 0.000312 0.000312 0.019195 0.890389
C(subj_group) 1.0 0.077314 0.077314 4.751328 0.034213
C(contraction_group):C(subj_group) 1.0 0.000423 0.000423 0.025979 0.872629
Residual 48.0 0.781058 0.016272 NaN NaN
P3i_cost_closeness_vitality_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.640778e+05 1.640778e+05 0.059177 0.808837
C(contraction_group) 1.0 1.832840e+07 1.832840e+07 6.610445 0.013293
C(subj_group) 1.0 7.708995e+05 7.708995e+05 0.278038 0.600419
C(contraction_group):C(subj_group) 1.0 9.042823e+06 9.042823e+06 3.261446 0.077199
Residual 48.0 1.330868e+08 2.772642e+06 NaN NaN
PO3i_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.030076 0.030076 10.695312 0.001992
C(contraction_group) 1.0 0.002371 0.002371 0.843089 0.363106
C(subj_group) 1.0 0.012640 0.012640 4.494936 0.039191
C(contraction_group):C(subj_group) 1.0 0.005047 0.005047 1.794858 0.186644
Residual 48.0 0.134981 0.002812 NaN NaN
PO3i_closeness_vitality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 6970.844053 6970.844053 1.615911 0.209789
C(contraction_group) 1.0 3534.539180 3534.539180 0.819341 0.369894
C(subj_group) 1.0 32806.565994 32806.565994 7.604889 0.008210
C(contraction_group):C(subj_group) 1.0 2110.534147 2110.534147 0.489243 0.487641
Residual 48.0 207066.173205 4313.878608 NaN NaN
PO3i_closeness_vitality_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 29534.648358 29534.648358 1.739265 0.193490
C(contraction_group) 1.0 12715.715763 12715.715763 0.748815 0.391156
C(subj_group) 1.0 134455.728571 134455.728571 7.917960 0.007075
C(contraction_group):C(subj_group) 1.0 10388.027789 10388.027789 0.611740 0.437975
Residual 48.0 815093.117975 16981.106624 NaN NaN
Fz_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.299252 1.299252 84.041609 4.027762e-12
C(contraction_group) 1.0 0.063177 0.063177 4.086597 4.881921e-02
C(subj_group) 1.0 0.001207 0.001207 0.078068 7.811330e-01
C(contraction_group):C(subj_group) 1.0 0.026289 0.026289 1.700507 1.984407e-01
Residual 48.0 0.742062 0.015460 NaN NaN
Fz_ecc_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 257.351819 257.351819 6.422757 0.014583
C(contraction_group) 1.0 173.211079 173.211079 4.322848 0.042970
C(subj_group) 1.0 5.170655 5.170655 0.129045 0.721000
C(contraction_group):C(subj_group) 1.0 29.302943 29.302943 0.731317 0.396705
Residual 48.0 1923.299708 40.068744 NaN NaN
Cz_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.024203 0.024203 20.598131 0.000038
C(contraction_group) 1.0 0.001353 0.001353 1.151857 0.288527
C(subj_group) 1.0 0.006294 0.006294 5.356732 0.024967
C(contraction_group):C(subj_group) 1.0 0.000386 0.000386 0.328127 0.569438
Residual 48.0 0.056400 0.001175 NaN NaN
Fp1c_degree_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.711750 1.711750 46.693748 1.324768e-08
C(contraction_group) 1.0 0.001174 0.001174 0.032026 8.587241e-01
C(subj_group) 1.0 0.216864 0.216864 5.915708 1.878566e-02
C(contraction_group):C(subj_group) 1.0 0.002373 0.002373 0.064736 8.002476e-01
Residual 48.0 1.759636 0.036659 NaN NaN
AF3c_degree_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 2.113271 2.113271 68.399088 8.621915e-11
C(contraction_group) 1.0 0.000188 0.000188 0.006080 9.381730e-01
C(subj_group) 1.0 0.149924 0.149924 4.852514 3.244000e-02
C(contraction_group):C(subj_group) 1.0 0.000012 0.000012 0.000372 9.846856e-01
Residual 48.0 1.483017 0.030896 NaN NaN
F3c_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.965383 1.965383 124.528360 6.190177e-15
C(contraction_group) 1.0 0.102972 0.102972 6.524371 1.386907e-02
C(subj_group) 1.0 0.102693 0.102693 6.506701 1.399057e-02
C(contraction_group):C(subj_group) 1.0 0.072602 0.072602 4.600110 3.705991e-02
Residual 48.0 0.757565 0.015783 NaN NaN
F3c_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.033159 0.033159 16.716094 0.000165
C(contraction_group) 1.0 0.013656 0.013656 6.884278 0.011624
C(subj_group) 1.0 0.014017 0.014017 7.066469 0.010638
C(contraction_group):C(subj_group) 1.0 0.013542 0.013542 6.826544 0.011956
Residual 48.0 0.095216 0.001984 NaN NaN
F7c_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.763794 1.763794 95.372152 5.469436e-13
C(contraction_group) 1.0 0.031690 0.031690 1.713553 1.967573e-01
C(subj_group) 1.0 0.082424 0.082424 4.456825 3.999587e-02
C(contraction_group):C(subj_group) 1.0 0.023140 0.023140 1.251204 2.688894e-01
Residual 48.0 0.887703 0.018494 NaN NaN
FC1c_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.003035 0.003035 1.875252 0.177248
C(contraction_group) 1.0 0.008066 0.008066 4.984031 0.030283
C(subj_group) 1.0 0.000205 0.000205 0.126723 0.723414
C(contraction_group):C(subj_group) 1.0 0.004249 0.004249 2.625783 0.111693
Residual 48.0 0.077677 0.001618 NaN NaN
FC5c_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.756319 1.756319 131.764599 2.292393e-15
C(contraction_group) 1.0 0.033259 0.033259 2.495183 1.207632e-01
C(subj_group) 1.0 0.094626 0.094626 7.099116 1.047100e-02
C(contraction_group):C(subj_group) 1.0 0.027957 0.027957 2.097401 1.540508e-01
Residual 48.0 0.639802 0.013329 NaN NaN
T8c_ecc_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 226.828940 226.828940 5.660437 0.021376
C(contraction_group) 1.0 173.399811 173.399811 4.327132 0.042872
C(subj_group) 1.0 1.639497 1.639497 0.040913 0.840561
C(contraction_group):C(subj_group) 1.0 30.427078 30.427078 0.759297 0.387886
Residual 48.0 1923.489043 40.072688 NaN NaN
CP5c_eigenvector_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.283669 0.283669 20.182861 0.000044
C(contraction_group) 1.0 0.044761 0.044761 3.184736 0.080650
C(subj_group) 1.0 0.059439 0.059439 4.229023 0.045197
C(contraction_group):C(subj_group) 1.0 0.015767 0.015767 1.121820 0.294828
Residual 48.0 0.674637 0.014055 NaN NaN
CP5c_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 1.657917 1.657917 105.571300 1.034954e-13
C(contraction_group) 1.0 0.033569 0.033569 2.137602 1.502431e-01
C(subj_group) 1.0 0.068404 0.068404 4.355770 4.221791e-02
C(contraction_group):C(subj_group) 1.0 0.021887 0.021887 1.393696 2.435990e-01
Residual 48.0 0.753804 0.015704 NaN NaN
CP5c_cost_eigenvector_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.283669 0.283669 20.182861 0.000044
C(contraction_group) 1.0 0.044761 0.044761 3.184736 0.080650
C(subj_group) 1.0 0.059439 0.059439 4.229023 0.045197
C(contraction_group):C(subj_group) 1.0 0.015767 0.015767 1.121820 0.294828
Residual 48.0 0.674637 0.014055 NaN NaN
PO3c_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.002448 0.002448 1.789698 0.187267
C(contraction_group) 1.0 0.008726 0.008726 6.379782 0.014897
C(subj_group) 1.0 0.000428 0.000428 0.312578 0.578702
C(contraction_group):C(subj_group) 1.0 0.009279 0.009279 6.783708 0.012210
Residual 48.0 0.065655 0.001368 NaN NaN
emg2_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.956229 0.956229 99.991839 2.536294e-13
C(contraction_group) 1.0 0.039652 0.039652 4.146394 4.726107e-02
C(subj_group) 1.0 0.015671 0.015671 1.638701 2.066560e-01
C(contraction_group):C(subj_group) 1.0 0.018804 0.018804 1.966349 1.672704e-01
Residual 48.0 0.459027 0.009563 NaN NaN
emg2_ecc_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 212.059072 212.059072 5.208442 0.026951
C(contraction_group) 1.0 172.115503 172.115503 4.227377 0.045237
C(subj_group) 1.0 0.225747 0.225747 0.005545 0.940952
C(contraction_group):C(subj_group) 1.0 24.764467 24.764467 0.608247 0.439277
Residual 48.0 1954.295615 40.714492 NaN NaN
emg2_cost_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.000481 0.000481 0.138172 0.711741
C(contraction_group) 1.0 0.014774 0.014774 4.243388 0.044848
C(subj_group) 1.0 0.000867 0.000867 0.249006 0.620055
C(contraction_group):C(subj_group) 1.0 0.008586 0.008586 2.466029 0.122901
Residual 48.0 0.167118 0.003482 NaN NaN
emg4_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.014861 0.014861 5.479129 0.023447
C(contraction_group) 1.0 0.029351 0.029351 10.821138 0.001884
C(subj_group) 1.0 0.005478 0.005478 2.019520 0.161753
C(contraction_group):C(subj_group) 1.0 0.023561 0.023561 8.686588 0.004936
Residual 48.0 0.130194 0.002712 NaN NaN
emg5_closeness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.941891 0.941891 80.915449 7.205280e-12
C(contraction_group) 1.0 0.048399 0.048399 4.157846 4.696890e-02
C(subj_group) 1.0 0.007642 0.007642 0.656542 4.217826e-01
C(contraction_group):C(subj_group) 1.0 0.022185 0.022185 1.905859 1.738192e-01
Residual 48.0 0.558741 0.011640 NaN NaN
emg5_closeness_vitality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 4.592700e+04 45927.000000 1.256838 0.267828
C(contraction_group) 1.0 1.510131e+05 151013.116318 4.132624 0.047615
C(subj_group) 1.0 2.151666e+03 2151.666129 0.058882 0.809305
C(contraction_group):C(subj_group) 1.0 2.282651e+04 22826.506831 0.624670 0.433205
Residual 48.0 1.754002e+06 36541.701904 NaN NaN
emg6_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.001624 0.001624 0.535466 0.467876
C(contraction_group) 1.0 0.022838 0.022838 7.531399 0.008503
C(subj_group) 1.0 0.001091 0.001091 0.359644 0.551525
C(contraction_group):C(subj_group) 1.0 0.008161 0.008161 2.691306 0.107435
Residual 48.0 0.145556 0.003032 NaN NaN
emg6_clusteringC_weight
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.005943 0.005943 4.045938 0.049911
C(contraction_group) 1.0 0.001474 0.001474 1.003489 0.321491
C(subj_group) 1.0 0.007909 0.007909 5.384082 0.024618
C(contraction_group):C(subj_group) 1.0 0.001040 0.001040 0.708016 0.404276
Residual 48.0 0.070511 0.001469 NaN NaN
emg6_cost_betweenness_centrality
df sum_sq mean_sq F PR(>F)
Intercept 1.0 0.001312 0.001312 0.340236 0.562424
C(contraction_group) 1.0 0.055301 0.055301 14.345932 0.000424
C(subj_group) 1.0 0.000494 0.000494 0.128023 0.722060
C(contraction_group):C(subj_group) 1.0 0.035802 0.035802 9.287605 0.003745
Residual 48.0 0.185030 0.003855 NaN NaN
Optional: ANOVA result and bp for a response variable
res_var = 'radius'
res.anova_stat(df=df, res_var=res_var, anova_model=res_var+' ~ C(contraction_group)*C(subj_group)',ss_typ=3)
print(res.anova_summary)
plt.figure()
ax = sns.boxplot(x="contraction_group", y=res_var, hue="subj_group", data=df, palette="Set3")
df sum_sq mean_sq F \
Intercept 1.0 51.571429 51.571429 137.869347
C(contraction_group) 1.0 0.285714 0.285714 0.763819
C(subj_group) 1.0 0.611914 0.611914 1.635872
C(contraction_group):C(subj_group) 1.0 0.002313 0.002313 0.006185
Residual 48.0 17.954887 0.374060 NaN
PR(>F)
Intercept 1.022717e-15
C(contraction_group) 3.864877e-01
C(subj_group) 2.070418e-01
C(contraction_group):C(subj_group) 9.376433e-01
Residual NaN

components
healthy vs patients - push
from scipy import stats
import seaborn as sns
import matplotlib.pyplot as plt
n_chs=40
# df = pd.read_csv(df_fName)
con_groupDiff_push_m_fName = os.path.join(global_results_dir,'iVC','con_groupDiff_push_m.eps') #patient - healthy subjects
alpha = 1e-2
t_stats = np.zeros((n_chs,n_chs))
p_values = np.zeros((n_chs,n_chs))
sig_cons_mask = np.zeros((n_chs,n_chs))
con_groupDiff_push = np.zeros((n_chs,n_chs))
con_groupDiff_push_dict = {}
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
SPMI_ch1ch2_patient_push = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
SPMI_ch1ch2_healthy_push = df.loc[(df['subj_group']=='healthy')&(df['contraction_group']=='push'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
t_stat,p_value = stats.ttest_ind(SPMI_ch1ch2_patient_push,SPMI_ch1ch2_healthy_push)
t_stats[ch1_idx][ch2_idx] = t_stat
p_values[ch1_idx][ch2_idx] = p_value
if p_value < alpha:
sig_cons_mask[ch1_idx][ch2_idx] = True
con_groupDiff_push[ch1_idx][ch2_idx] = cohend(SPMI_ch1ch2_patient_push, SPMI_ch1ch2_healthy_push)
con_groupDiff_push_dict['SPMI_'+ch1_name+'_'+ch2_name] = [SPMI_ch1ch2_patient_push, SPMI_ch1ch2_healthy_push]
with sns.axes_style("white"):
fig, ax = plt.subplots(figsize=(9, 9))
ax = sns.heatmap(con_groupDiff_push, center = 0, square=True,cmap="bwr")
figure = ax.get_figure()
figure.savefig(con_groupDiff_push_m_fName)

# graph
########### customization ###################
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_lParetic = ['Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'Fz','Cz','Pz','Oz',
'Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_rParetic = ['Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'Fz','Cz','Pz','Oz',
'Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
EMGs=[EMGChannel(2,150,'emg1'),EMGChannel(2,170,'emg2'),EMGChannel(2, 240,'emg3'),EMGChannel(2,350,'emg4'),
EMGChannel(2, 80,'emg5'),EMGChannel(4,140,'emg6'),EMGChannel(4, 310,'emg7'),
EMGChannel(4,90,'emg8')]
EEGs = [EEGChannel(x=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][0]*50,
y=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][1]*50,
name=ch) for ch in chs_list_image]
con_groupDiff_push_g_fName = os.path.join(global_results_dir,'con_groupDiff_push_g.eps') #after-before
#########################################
fig,ax = plt.subplots(figsize=(16, 16))
ax = plt.gca()
ax = plotTopoEMG(EMGs,ax)
ax = plotTopoEEG(EEGs,ax)
fig,ax = graph_plot_fixMapper(con_groupDiff_push,con_groupDiff_push,fig,ax)
plt.xlim([-12,12])
plt.ylim([-12,12])
fig.savefig(con_groupDiff_push_g_fName)

#bp to see the distribution
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from statannot import add_stat_annotation
# df = None
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
dfs2concate = []
con_sig_name=[]
fig,ax = plt.subplots(figsize=(20, 12))
dfs2bp = []
for con_groupDiff in con_groupDiff_push_dict.keys():
df2bp = np.concatenate((df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'), con_groupDiff].to_numpy().astype(np.float),
df.loc[(df['subj_group']=='healthy')&(df['contraction_group']=='push'), con_groupDiff].to_numpy().astype(np.float)))
order = np.concatenate((np.array(['patient']*len(patient_subjs)),np.array(['healthy']*len(healthy_subjs))))
data = {'con_name':con_groupDiff,'SPMI':df2bp,'order':order}
dfs2bp.append(pd.DataFrame(data,columns = ['con_name','SPMI','order']))
df_bp = pd.concat(dfs2bp)
y = 'SPMI'
hue = 'order'
ax = sns.boxplot(data=df_bp, x='con_name', y=y, hue=hue,ax=ax,width = 0.4)
add_stat_annotation(ax,data=df_bp, x='con_name', y=y,
box_pairs=[((i,"patient"),(i,"healthy"))
for i in con_groupDiff_push_dict.keys()],
hue=hue,test='t-test_ind',text_format='full',fontsize=24,verbose=1)
plt.legend(loc='upper left', bbox_to_anchor = (0.7,1),prop={'size': 24})
# con_diff_prePostTMS_bp_fName = os.path.join(global_results_dir,'prePostTMS_sigConnection_bp.eps')
# plt.savefig(con_diff_prePostTMS_bp_fName)
# plt.cla()
SPMI_F7c_FC1c_patient v.s. SPMI_F7c_FC1c_healthy: t-test independent samples with Bonferroni correction, P_val=2.578e-02 stat=3.122e+00
SPMI_Pz_O1c_patient v.s. SPMI_Pz_O1c_healthy: t-test independent samples with Bonferroni correction, P_val=3.215e-02 stat=-3.026e+00
SPMI_T8c_emg1_patient v.s. SPMI_T8c_emg1_healthy: t-test independent samples with Bonferroni correction, P_val=5.612e-03 stat=3.771e+00
SPMI_P3c_emg3_patient v.s. SPMI_P3c_emg3_healthy: t-test independent samples with Bonferroni correction, P_val=5.477e-03 stat=3.781e+00
SPMI_PO3c_emg2_patient v.s. SPMI_PO3c_emg2_healthy: t-test independent samples with Bonferroni correction, P_val=8.784e-03 stat=3.582e+00
<matplotlib.legend.Legend at 0x238ba93f9b0>

regression-push
import scipy
from numpy import mean
from numpy import var
from math import sqrt
import statsmodels.api as sm
alpha = 1e-2
r2_threshold = 0.5
related_cons=[]
model_stats = []
FMAs = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'),'FMA'].to_numpy().astype(np.float)
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
SPMI_ch1ch2_patient_push = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
SPMI_ch1ch2_patient_push =sm.add_constant(SPMI_ch1ch2_patient_push)
model = sm.OLS(FMAs,SPMI_ch1ch2_patient_push).fit()
if (model.f_pvalue<alpha) & (model.rsquared_adj>0.5):
print(model.summary())
print('SPMI_'+ch1_name+'_'+ch2_name)
related_cons.append([ch1_name,ch2_name])
model_stats.append([round(model.f_pvalue,2),round(model.rsquared_adj,2)])
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.618
Model: OLS Adj. R-squared: 0.591
Method: Least Squares F-statistic: 22.63
Date: Tue, 30 Mar 2021 Prob (F-statistic): 0.000306
Time: 21:57:06 Log-Likelihood: -55.995
No. Observations: 16 AIC: 116.0
Df Residuals: 14 BIC: 117.5
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 47.4290 3.247 14.605 0.000 40.464 54.394
x1 -2.5081 0.527 -4.757 0.000 -3.639 -1.377
==============================================================================
Omnibus: 1.129 Durbin-Watson: 2.509
Prob(Omnibus): 0.569 Jarque-Bera (JB): 0.376
Skew: 0.375 Prob(JB): 0.828
Kurtosis: 3.054 Cond. No. 9.48
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_C3i_F3c
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.608
Model: OLS Adj. R-squared: 0.580
Method: Least Squares F-statistic: 21.73
Date: Tue, 30 Mar 2021 Prob (F-statistic): 0.000367
Time: 21:57:06 Log-Likelihood: -56.194
No. Observations: 16 AIC: 116.4
Df Residuals: 14 BIC: 117.9
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 49.1392 3.588 13.697 0.000 41.445 56.834
x1 -2.4626 0.528 -4.662 0.000 -3.595 -1.330
==============================================================================
Omnibus: 1.403 Durbin-Watson: 1.888
Prob(Omnibus): 0.496 Jarque-Bera (JB): 1.079
Skew: 0.421 Prob(JB): 0.583
Kurtosis: 2.045 Cond. No. 11.4
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_CP5i_F3c
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.597
Model: OLS Adj. R-squared: 0.568
Method: Least Squares F-statistic: 20.72
Date: Tue, 30 Mar 2021 Prob (F-statistic): 0.000452
Time: 21:57:06 Log-Likelihood: -56.423
No. Observations: 16 AIC: 116.8
Df Residuals: 14 BIC: 118.4
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 49.0124 3.639 13.468 0.000 41.207 56.818
x1 -2.3732 0.521 -4.552 0.000 -3.491 -1.255
==============================================================================
Omnibus: 0.462 Durbin-Watson: 2.398
Prob(Omnibus): 0.794 Jarque-Bera (JB): 0.533
Skew: 0.067 Prob(JB): 0.766
Kurtosis: 2.116 Cond. No. 11.7
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_P3i_F3c
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.617
Model: OLS Adj. R-squared: 0.590
Method: Least Squares F-statistic: 22.57
Date: Tue, 30 Mar 2021 Prob (F-statistic): 0.000310
Time: 21:57:06 Log-Likelihood: -56.008
No. Observations: 16 AIC: 116.0
Df Residuals: 14 BIC: 117.6
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 57.0325 4.954 11.513 0.000 46.408 67.657
x1 -6.4677 1.361 -4.751 0.000 -9.387 -3.548
==============================================================================
Omnibus: 1.528 Durbin-Watson: 2.735
Prob(Omnibus): 0.466 Jarque-Bera (JB): 0.714
Skew: 0.517 Prob(JB): 0.700
Kurtosis: 2.979 Cond. No. 8.94
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_F3c_C3c
#visualization
import seaborn as sns
FMAs = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'),'FMA'].to_numpy().astype(np.float)
for i in range(len(related_cons)):
ch1_name = related_cons[i][0]
ch2_name = related_cons[i][1]
SPMI_ch1ch2_patient_push = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
plt.figure()
ax = sns.regplot(x=SPMI_ch1ch2_patient_push, y=FMAs)
ax.set(xlabel=ch1_name+'_'+ch2_name, ylabel='FMA')
ax.set_xlim([-0.1,16])
ax.set_ylim([5,60])
plt.text(2, 10, "p_F="+str(model_stats[i][0])+", r2_adj="+str(model_stats[i][1]), horizontalalignment='left',
size='medium', color='black', weight='semibold')
regPlot_fName = os.path.join(global_results_dir,'iVC',ch1_name+'-'+ch2_name+'push_FMAreg.eps')
figure = ax.get_figure()
figure.savefig(regPlot_fName)
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.




# graph plot
# graph
########### customization ###################
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_lParetic = ['Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'Fz','Cz','Pz','Oz',
'Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_rParetic = ['Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'Fz','Cz','Pz','Oz',
'Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
EMGs=[EMGChannel(2,150,'emg1'),EMGChannel(2,170,'emg2'),EMGChannel(2, 240,'emg3'),EMGChannel(2,350,'emg4'),
EMGChannel(2, 80,'emg5'),EMGChannel(4,140,'emg6'),EMGChannel(4, 310,'emg7'),
EMGChannel(4,90,'emg8')]
EEGs = [EEGChannel(x=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][0]*50,
y=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][1]*50,
name=ch) for ch in chs_list_image]
con_reg_g_fName = os.path.join(global_results_dir,'con_reg_g.eps') #after-before
#########################################
con_reg = np.zeros([n_chs,n_chs])
for con in related_cons:
con_reg[chs_list_image.index(con[0]),chs_list_image.index(con[1])]=1
fig,ax = plt.subplots(figsize=(18, 16))
ax = plt.gca()
ax = plotTopoEMG(EMGs,ax)
ax = plotTopoEEG(EEGs,ax)
fig,ax = graph_plot_fixMapper(con_reg,con_reg,fig,ax)
plt.xlim([-12,12])
plt.ylim([-12,12])
fig.savefig(con_reg_g_fName)

# integral model
X=[]
for con in related_cons:
X.append(df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='push'),
'SPMI_'+con[0]+'_'+con[1]].to_numpy().astype(np.float))
X=np.array(X)
X=np.swapaxes(X,0,1)
X=sm.add_constant(X)
model = sm.OLS(FMAs,X).fit()
predictions = model.predict(X)
print(model.summary())
print('\n')
print(predictions)
print(FMAs)
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.753
Model: OLS Adj. R-squared: 0.663
Method: Least Squares F-statistic: 8.393
Date: Tue, 30 Mar 2021 Prob (F-statistic): 0.00234
Time: 22:15:34 Log-Likelihood: -52.496
No. Observations: 16 AIC: 115.0
Df Residuals: 11 BIC: 118.9
Df Model: 4
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 55.6930 4.571 12.183 0.000 45.632 65.754
x1 -0.9026 1.125 -0.802 0.439 -3.379 1.574
x2 -0.8519 0.951 -0.896 0.390 -2.945 1.241
x3 0.0233 1.153 0.020 0.984 -2.515 2.561
x4 -3.4196 1.814 -1.885 0.086 -7.412 0.573
==============================================================================
Omnibus: 4.940 Durbin-Watson: 2.779
Prob(Omnibus): 0.085 Jarque-Bera (JB): 2.375
Skew: 0.829 Prob(JB): 0.305
Kurtosis: 3.903 Cond. No. 29.0
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[19.3967171 49.00864414 17.73351263 39.46690484 26.36668896 12.31203864
41.0883702 47.96277939 41.79670717 36.74481556 37.07756046 32.32217292
42.81885559 32.65025043 50.34512571 45.90885626]
[20. 47. 20. 37. 19. 15. 58. 39. 51. 31. 38. 23. 42. 35. 53. 45.]
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
healthy vs patients - pull
from scipy import stats
import seaborn as sns
import matplotlib.pyplot as plt
n_chs=40
# df = pd.read_csv(df_fName)
con_groupDiff_pull_m_fName = os.path.join(global_results_dir,'iVC','con_groupDiff_pull_m.eps') #patient - healthy subjects
alpha = 1e-2
t_stats = np.zeros((n_chs,n_chs))
p_values = np.zeros((n_chs,n_chs))
sig_cons_mask = np.zeros((n_chs,n_chs))
con_groupDiff_pull = np.zeros((n_chs,n_chs))
con_groupDiff_pull_dict = {}
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
SPMI_ch1ch2_patient_pull = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='pull'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
SPMI_ch1ch2_healthy_pull = df.loc[(df['subj_group']=='healthy')&(df['contraction_group']=='pull'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
t_stat,p_value = stats.ttest_ind(SPMI_ch1ch2_patient_pull,SPMI_ch1ch2_healthy_pull)
t_stats[ch1_idx][ch2_idx] = t_stat
p_values[ch1_idx][ch2_idx] = p_value
if p_value < alpha:
sig_cons_mask[ch1_idx][ch2_idx] = True
con_groupDiff_pull[ch1_idx][ch2_idx] = cohend(SPMI_ch1ch2_patient_pull, SPMI_ch1ch2_healthy_pull)
con_groupDiff_pull_dict['SPMI_'+ch1_name+'_'+ch2_name] = [SPMI_ch1ch2_patient_pull, SPMI_ch1ch2_healthy_pull]
with sns.axes_style("white"):
fig, ax = plt.subplots(figsize=(9, 9))
ax = sns.heatmap(con_groupDiff_pull, center = 0, square=True,cmap="bwr")
figure = ax.get_figure()
figure.savefig(con_groupDiff_pull_m_fName)

# graph
########### customization ###################
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_lParetic = ['Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'Fz','Cz','Pz','Oz',
'Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_rParetic = ['Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'Fz','Cz','Pz','Oz',
'Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
EMGs=[EMGChannel(2,150,'emg1'),EMGChannel(2,170,'emg2'),EMGChannel(2, 240,'emg3'),EMGChannel(2,350,'emg4'),
EMGChannel(2, 80,'emg5'),EMGChannel(4,140,'emg6'),EMGChannel(4, 310,'emg7'),
EMGChannel(4,90,'emg8')]
EEGs = [EEGChannel(x=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][0]*50,
y=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][1]*50,
name=ch) for ch in chs_list_image]
con_groupDiff_pull_g_fName = os.path.join(global_results_dir,'con_groupDiff_pull_g.eps') #after-before
#########################################
fig,ax = plt.subplots(figsize=(18, 16))
ax = plt.gca()
ax = plotTopoEMG(EMGs,ax)
ax = plotTopoEEG(EEGs,ax)
fig,ax = graph_plot_fixMapper(con_groupDiff_pull,con_groupDiff_pull,fig,ax)
plt.xlim([-12,12])
plt.ylim([-12,12])
fig.savefig(con_groupDiff_pull_g_fName)

#bp to see the distribution, the annotated pvalues are
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from statannot import add_stat_annotation
# df = None
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
fig,ax = plt.subplots(figsize=(22, 10))
dfs2bp = []
for con_groupDiff in con_groupDiff_pull_dict.keys():
df2bp = np.concatenate((df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='pull'), con_groupDiff].to_numpy().astype(np.float),
df.loc[(df['subj_group']=='healthy')&(df['contraction_group']=='pull'), con_groupDiff].to_numpy().astype(np.float)))
order = np.concatenate((np.array(['patient']*len(patient_subjs)),np.array(['healthy']*len(healthy_subjs))))
data = {'con_name':con_groupDiff,'SPMI':df2bp,'order':order}
dfs2bp.append(pd.DataFrame(data,columns = ['con_name','SPMI','order']))
df_bp = pd.concat(dfs2bp)
y = 'SPMI'
hue = 'order'
ax = sns.boxplot(data=df_bp, x='con_name', y=y, hue=hue,ax=ax,width = 0.4)
add_stat_annotation(ax,data=df_bp, x='con_name', y=y,
box_pairs=[((i,"patient"),(i,"healthy"))
for i in con_groupDiff_pull_dict.keys()],
hue=hue,test='t-test_ind',text_format='full',fontsize=24,verbose=1)
plt.legend(loc='upper left', bbox_to_anchor = (0.7,1),prop={'size': 24})
# con_diff_prePostTMS_bp_fName = os.path.join(global_results_dir,'prePostTMS_sigConnection_bp.eps')
# plt.savefig(con_diff_prePostTMS_bp_fName)
# plt.cla()
SPMI_PO3c_O1c_patient v.s. SPMI_PO3c_O1c_healthy: t-test independent samples with Bonferroni correction, P_val=8.051e-02 stat=-3.007e+00
SPMI_emg3_emg6_patient v.s. SPMI_emg3_emg6_healthy: t-test independent samples with Bonferroni correction, P_val=7.847e-02 stat=3.019e+00
SPMI_emg5_emg6_patient v.s. SPMI_emg5_emg6_healthy: t-test independent samples with Bonferroni correction, P_val=1.091e-01 stat=-2.874e+00
SPMI_CP1c_emg1_patient v.s. SPMI_CP1c_emg1_healthy: t-test independent samples with Bonferroni correction, P_val=1.069e-01 stat=2.883e+00
SPMI_C3i_CP5i_patient v.s. SPMI_C3i_CP5i_healthy: t-test independent samples with Bonferroni correction, P_val=7.997e-02 stat=-3.010e+00
SPMI_FC5i_emg1_patient v.s. SPMI_FC5i_emg1_healthy: t-test independent samples with Bonferroni correction, P_val=8.455e-02 stat=2.986e+00
SPMI_T7i_emg3_patient v.s. SPMI_T7i_emg3_healthy: t-test independent samples with Bonferroni correction, P_val=9.412e-02 stat=2.939e+00
SPMI_CP1i_emg2_patient v.s. SPMI_CP1i_emg2_healthy: t-test independent samples with Bonferroni correction, P_val=1.112e-01 stat=2.865e+00
SPMI_CP5i_P3i_patient v.s. SPMI_CP5i_P3i_healthy: t-test independent samples with Bonferroni correction, P_val=6.083e-02 stat=-3.129e+00
SPMI_Cz_emg3_patient v.s. SPMI_Cz_emg3_healthy: t-test independent samples with Bonferroni correction, P_val=6.348e-03 stat=4.086e+00
SPMI_Pz_O1c_patient v.s. SPMI_Pz_O1c_healthy: t-test independent samples with Bonferroni correction, P_val=4.948e-02 stat=-3.218e+00
SPMI_F3c_emg2_patient v.s. SPMI_F3c_emg2_healthy: t-test independent samples with Bonferroni correction, P_val=5.214e-02 stat=-3.196e+00
<matplotlib.legend.Legend at 0x238ba0d6a20>

reg-pull
import scipy
from numpy import mean
from numpy import var
from math import sqrt
import statsmodels.api as sm
alpha = 1e-2
r2_threshold = 0.5
related_cons=[]
model_stats = []
FMAs = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='pull'),'FMA'].to_numpy().astype(np.float)
for ch1_idx in range(n_chs):
for ch2_idx in np.arange(ch1_idx+1,n_chs):
ch1_name = chs_list_image[ch1_idx]
ch2_name = chs_list_image[ch2_idx]
SPMI_ch1ch2_patient_pull = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='pull'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
SPMI_ch1ch2_patient_pull =sm.add_constant(SPMI_ch1ch2_patient_pull)
model = sm.OLS(FMAs,SPMI_ch1ch2_patient_pull).fit()
if (model.f_pvalue<alpha) & (model.rsquared_adj>0.5):
print(model.summary())
print('SPMI_'+ch1_name+'_'+ch2_name)
related_cons.append([ch1_name,ch2_name,model.params[1]])
sciNotation = '%E' % model.f_pvalue
roundedSciNotation = str(round(float(sciNotation.split('E')[0].rstrip('0').rstrip('.')),2)) + 'E' + sciNotation.split('E')[1]
model_stats.append([roundedSciNotation,round(model.rsquared_adj,2)])
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.542
Model: OLS Adj. R-squared: 0.510
Method: Least Squares F-statistic: 16.59
Date: Thu, 01 Apr 2021 Prob (F-statistic): 0.00114
Time: 00:16:46 Log-Likelihood: -57.437
No. Observations: 16 AIC: 118.9
Df Residuals: 14 BIC: 120.4
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 46.7539 3.564 13.117 0.000 39.109 54.399
x1 -2.3027 0.565 -4.073 0.001 -3.515 -1.090
==============================================================================
Omnibus: 0.060 Durbin-Watson: 2.682
Prob(Omnibus): 0.971 Jarque-Bera (JB): 0.288
Skew: 0.029 Prob(JB): 0.866
Kurtosis: 2.345 Cond. No. 9.73
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_C3i_Fp1c
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.537
Model: OLS Adj. R-squared: 0.504
Method: Least Squares F-statistic: 16.25
Date: Thu, 01 Apr 2021 Prob (F-statistic): 0.00124
Time: 00:16:46 Log-Likelihood: -57.527
No. Observations: 16 AIC: 119.1
Df Residuals: 14 BIC: 120.6
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 46.3280 3.515 13.180 0.000 38.789 53.867
x1 -2.1892 0.543 -4.031 0.001 -3.354 -1.024
==============================================================================
Omnibus: 0.712 Durbin-Watson: 2.413
Prob(Omnibus): 0.700 Jarque-Bera (JB): 0.296
Skew: 0.326 Prob(JB): 0.862
Kurtosis: 2.863 Cond. No. 9.78
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_C3i_F3c
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.576
Model: OLS Adj. R-squared: 0.545
Method: Least Squares F-statistic: 19.00
Date: Thu, 01 Apr 2021 Prob (F-statistic): 0.000654
Time: 00:16:46 Log-Likelihood: -56.830
No. Observations: 16 AIC: 117.7
Df Residuals: 14 BIC: 119.2
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 47.8130 3.559 13.434 0.000 40.180 55.446
x1 -2.3281 0.534 -4.359 0.001 -3.474 -1.183
==============================================================================
Omnibus: 2.953 Durbin-Watson: 2.202
Prob(Omnibus): 0.228 Jarque-Bera (JB): 1.620
Skew: 0.779 Prob(JB): 0.445
Kurtosis: 3.069 Cond. No. 10.7
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_CP5i_F3c
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.551
Model: OLS Adj. R-squared: 0.519
Method: Least Squares F-statistic: 17.20
Date: Thu, 01 Apr 2021 Prob (F-statistic): 0.000987
Time: 00:16:46 Log-Likelihood: -57.279
No. Observations: 16 AIC: 118.6
Df Residuals: 14 BIC: 120.1
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 46.8086 3.523 13.287 0.000 39.253 54.365
x1 -1.9979 0.482 -4.147 0.001 -3.031 -0.965
==============================================================================
Omnibus: 0.600 Durbin-Watson: 2.575
Prob(Omnibus): 0.741 Jarque-Bera (JB): 0.450
Skew: 0.365 Prob(JB): 0.798
Kurtosis: 2.622 Cond. No. 11.2
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_P3i_F3c
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
C:\Users\aegea\Anaconda3\envs\p3.7\lib\site-packages\scipy\stats\stats.py:1604: UserWarning: kurtosistest only valid for n>=20 ... continuing anyway, n=16
"anyway, n=%i" % int(n))
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.538
Model: OLS Adj. R-squared: 0.505
Method: Least Squares F-statistic: 16.31
Date: Thu, 01 Apr 2021 Prob (F-statistic): 0.00122
Time: 00:16:47 Log-Likelihood: -57.509
No. Observations: 16 AIC: 119.0
Df Residuals: 14 BIC: 120.6
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 45.1462 3.298 13.688 0.000 38.072 52.220
x1 -7.7851 1.927 -4.039 0.001 -11.919 -3.651
==============================================================================
Omnibus: 2.817 Durbin-Watson: 1.630
Prob(Omnibus): 0.245 Jarque-Bera (JB): 1.364
Skew: 0.362 Prob(JB): 0.506
Kurtosis: 1.766 Cond. No. 2.87
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
SPMI_Cz_emg6
#visualization
import seaborn as sns
FMAs = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='pull'),'FMA'].to_numpy().astype(np.float)
for i in range(len(related_cons)):
ch1_name = related_cons[i][0]
ch2_name = related_cons[i][1]
SPMI_ch1ch2_patient_pull = df.loc[(df['subj_group']=='patient')&(df['contraction_group']=='pull'),
'SPMI_'+ch1_name+'_'+ch2_name].to_numpy().astype(np.float)
plt.figure()
ax = sns.regplot(x=SPMI_ch1ch2_patient_pull, y=FMAs)
ax.set(xlabel=ch1_name+'_'+ch2_name, ylabel='FMA')
ax.set_xlim([-0.5,16])
ax.set_ylim([5,60])
plt.text(2, 10, "p_F="+str(model_stats[i][0])+", r2_adj="+str(model_stats[i][1]), horizontalalignment='left',
size='medium', color='black', weight='semibold')
regPlot_fName = os.path.join(global_results_dir,'iVC',ch1_name+'-'+ch2_name+'pull_FMAreg.eps')
figure = ax.get_figure()
figure.savefig(regPlot_fName)
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.





# graph plot
# graph
########### customization ###################
chs_list_image = ['Fp1i','AF3i','F3i','F7i','FC1i','FC5i','C3i','T7i','CP1i','CP5i','P3i','P7i','PO3i','O1i',
'Fz','Cz','Pz','Oz',
'Fp1c','AF3c','F3c','F7c','FC1c','FC5c','C3c','T8c','CP1c','CP5c','P3c','P7c','PO3c','O1c',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_lParetic = ['Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'Fz','Cz','Pz','Oz',
'Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
chs_list_preImage_rParetic = ['Fp2','AF4','F4','F8','FC2','FC6','C4','T8','CP2','CP6','P4','P8','PO4','O2',
'Fz','Cz','Pz','Oz',
'Fp1','AF3','F3','F7','FC1','FC5','C3','T7','CP1','CP5','P3','P7','PO3','O1',
'emg1','emg2','emg3','emg4','emg5','emg6','emg7','emg8']
EMGs=[EMGChannel(2,150,'emg1'),EMGChannel(2,170,'emg2'),EMGChannel(2, 240,'emg3'),EMGChannel(2,350,'emg4'),
EMGChannel(2, 80,'emg5'),EMGChannel(4,140,'emg6'),EMGChannel(4, 310,'emg7'),
EMGChannel(4,90,'emg8')]
EEGs = [EEGChannel(x=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][0]*50,
y=epochs_hybrid.info['chs'][epochs_hybrid.info['ch_names'].index(chs_list_preImage_rParetic[chs_list_image.index(ch)])]['loc'][1]*50,
name=ch) for ch in chs_list_image]
con_reg_pull_g_fName = os.path.join(global_results_dir,'con_reg_pull_g.eps') #after-before
#########################################
con_reg = np.zeros([n_chs,n_chs])
for con in related_cons:
con_reg[chs_list_image.index(con[0]),chs_list_image.index(con[1])]= con[2]
fig,ax = plt.subplots(figsize=(18, 16))
ax = plt.gca()
ax = plotTopoEMG(EMGs,ax)
ax = plotTopoEEG(EEGs,ax)
fig,ax = graph_plot_fixMapper(con_reg,con_reg,fig,ax)
plt.xlim([-12,12])
plt.ylim([-12,12])
fig.savefig(con_reg_pull_g_fName)

